Introduction
The world today is riding the crest of a wave of advancing information in Molecular Biology. Information is being generated so fast that it is impossible for researchers in the field to keep up with the data flow, and the interpretation lags far behind the generation of data. Our understanding of the complexities of function and structure inside the cell is revolutionized again and again as we uncover new details of cellular processes.
In this context it is a particular tragedy that we blindly continued to dogmatize a fundamental theory in biology that is over 150 years old - the theory of naturalistic evolution. It was understandable for Darwin and other early protagonists of naturalistic evolution to discount the difficulties involved in the evolution of complex forms when virtually nothing was known about them. This was the case when Darwin first began formalizing his version of a theory for the spontaneous and undirected evolution of living forms in the first half of the 19th century.
It remained so over the subsequent century. But during the past 30 years, the tools of modern molecular systematics, along with the advances in our understanding of cellular and molecular processes in a wide spectrum of organisms have changed that picture. It is now possible to do detailed comparisons of the molecular features of a variety of organisms and to construct phylogenetic linkages between organisms based upon these comparisons. With such powerful tools available, it is no longer necessary to guess what kinds of processes were operative in organisms that are no longer available for study. Much of the molecular architecture of such forms can be reconstructed from data readily available to us today. The conclusions of such work are rather surprising and comprise the topic of this presentation.

Figure 1. The Devonian trilobite Phacops moroccensis from the Atlas Mountains in Morocco. This large and spectacular trilobite has complex features such as compound (schizochroal, in this case) eyes, swimmerets, compound jointed appendages, antennae, cephalized body form, compound mouth parts, and othercomplexities in common with modern insects and with the first trilobites found in the Lower Cambrian worldwide. What can we know about the molecular biology of this extinct form?
Trilobites are extinct members of the phylum Arthropoda, to which the modern insects belong. These creatures left a long and detailed fossil record in rocks beginning in earliest Cambrian and ending in the Permian. Trilobites were exquisite forms having elaborate segmented bodies, cephalized nervous systems, with jointed appendages and swimmerets, antennae and compound eyes. Because trilobites are extinct forms, we know very little of their life habits except for what we can deduce from association with other forms that do have living representatives, and from careful reconstructions of the depositional environments in which they are found.
However, the theory of evolution has provided us a mechanism for reconstructing in unimagined detail, the physiological and molecular biological nature of this first widely distributed metazoan form. The reconstruction is of great significance in providing us with a picture of the richness and complexity of the earliest pervasive metazoan creatures. It will also contribute substantially to our understanding of the processes that would have had to precede the appearance of these amazing creatures that nearly everywhere mark the boundary between rocks essentially barren of metazoan life, and those rocks containing abundant evidence of such life. Before we begin to explore the nature of the trilobite, let us lay some fundamental groundwork for the premises we will exploit in our reconstruction. I will
- demonstrate using a fundamental assumption of evolutionary theory, that we can know in exquisite detail, the molecular biology of the trilobite;
- show that trilobites are every bit as complex as any modern form at the molecular level;
and, in the absence of any physical evidence for evolution of complex systems, or for the increase in information content of existing complex systems,
- show that to belief in the theory of evolution is a matter of pure faith, since there is no physical evidence for increase in information content of any complex system. Another theory of explanation of origins - Special Creation - has scientific precedence because it does offer an explanation for origins that comports with the data.
Origin of Cells
All living organisms, including trilobites, are composed of cells. The theory of evolution proposes that these cells arose in the distant past from one or more simple living systems derived by natural processes from materials present on the prebiotic earth. These primitive protocells became established and over vast periods of time developed complex systems capable of efficient replication of the components necessary for life.
During this time, the details of the genetic code were worked out, the systems of enzymes necessary for replicating the DNA were perfected, the enzymes required to produce functional messenger RNAs developed, and the apparatus for producing proteins from the information contained in the messenger RNA was established. Whether this present system was the first developed, or whether a much simpler system involving only complex molecules of RNA capable of self-replication and catalytic activity preceded it, is an area of active speculation today.
This latter suggestion at first appeared to offer a way out of the dilemma posed by the requirement for the simultaneous appearance of proteins and DNA to code for those same proteins. But at present there is little evidence of a broadly significant role for these catalytic RNA molecules or "ribozymes," in modern cells, and the problem of changing over from a system of ribozymes to one of proteins governed by DNA remains enigmatic. Although the origin of life is not the subject of this paper, it is worth noting that this scenario, or any other scenario, for the spontaneous origin of a living cell belongs in the realm of science fiction. In any case, it is abundantly clear that before earliest Cambrian, the details of the modern eukaryotic cell of which trilobites were composed were fully accomplished, as we will see.
Revealing the Past
What can we know about the molecular and cellular biology and physiology of the trilobite? The premise underlying this presentation is that we can determine in exquisite and precise detail the mechanisms that were operating in the cells and tissues of the trilobite. This premise is grounded in a fundamental construct of the evolutionary theory: that traits shared in common by disparate organisms at the molecular or cellular level require a shared evolutionary ancestry.
This assumption is widely accepted and undergirds the entire evolutionary enterprise, being the basis of all modern evolutionary taxonomy. Although some anatomical similarities are considered to be examples of convergent (independently derived and not genetically related) evolution, such as flight in insects, reptiles, birds and mammals, such cases are easily identified, and similarities that exist at the cellular and molecular level are generally considered to indicate a common ancestry.
Thus, molecular features shared by the common garden pea and man would require there to have been at some time in the distant past, a common ancestor possessing those common features (figure 2). Any other conclusion would require highly unlikely events to have been replicated repeatedly with exacting precision, falsifying the fundamental assumption of molecular systematics and taxing credulity beyond limits.
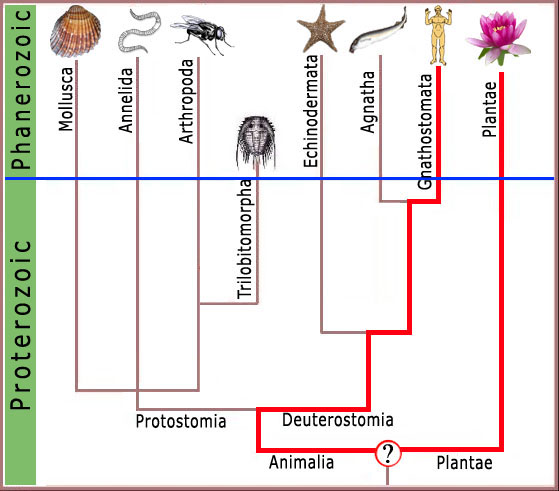
Figure 2. A chart representing standard geologic time on the y axis and showing hypothetical ranges for various groups of animals and plants on the x axis. The thin blue line represents the division between data (above the line) and speculation in the absence of data (below the line). Note that any molecular biological feature in common between plants and animals must have been present in the hypothetical last common ancestor (LCA) of plants and animals.
Consequently, any complex feature shared by modern arthropods and man (figure 3), or arthropods and the garden pea (figure 4), must have been present in the ancestor common to both forms.
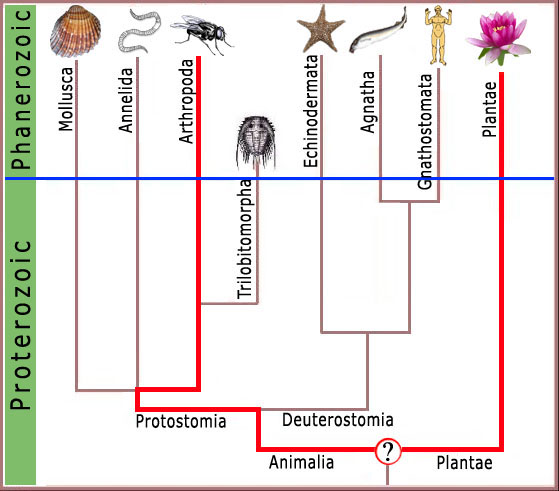
Figure 3. LCA of plant and arthropod (insect alliance) would have been deep in the Precambrian where no complex features at the molecular level would be expected.
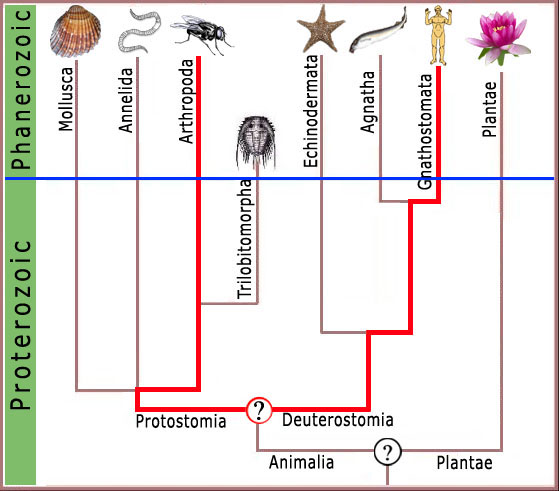
Figure 4. Likewise LCA for human and arthropod would be expected to be a featureless blob deep in the Precambrian with no complex features of modern organisms.
Thus, the presence of features of cellular or molecular biology in common between modern arthropods and man or other modern forms, requires that these features were shared by the common ancestor of arthropods and man. Since trilobites were arthropods, they too must have exhibited these features, and we can attribute these complex features to this early metazoan with confidence.
We will look at several of a large potential number of complex molecular biological systems. It will, of course, be necessary to include some material of a rather technical nature in order to establish the level of complexity present in cells. This is unavoidable, because this background is needed to develop the salient points. These details are well known to molecular biologists, but it is not necessary to be a molecular biologist or to understand the details of the complexity in order to understand the significance of the arguments. I will now begin to consider a few of the complex features of the trilobite.
The Eukaryotic Chromosome
The eukaryotic cell that comprises all of the organisms we are generally familiar with, including humans, carries the vast array of information it contains coded in the form of long (less than 1cm to 15 cm or more) molecules of DNA. Every somatic cell in the human body has a complement of 46 of these molecules. All the DNA of a single human cell would extend to nearly two meters if the DNA molecules from all 46 chromosomes were placed end to end.
This DNA is housed in a nucleus with a diameter of about 10 micrometers. So, the length of DNA in the nucleus of a single human cell is 200,000 times the radius of the nucleus. An equivalent illustration would be to have 70 kilometers of kite string in a shoebox! How can the cell cope? In order to divide, it must replicate the entire length of each chromosome, making nearly 4 meters of DNA. Then it must divide that DNA neatly between the two resulting daughter cells. To expedite this process, the DNA is separated into individual chromosomes averaging about 50 mm of DNA each in humans (figure 5).
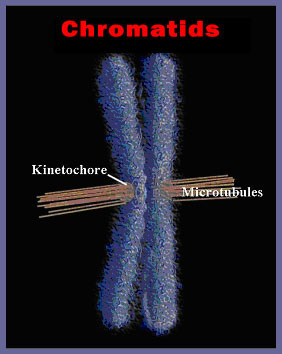
Figure 5. A single human chromosome, here shown doubled just prior to division in the spindle apparatus of the dividing cell. The microtubules have attached to the kinetochore and oriented the chromosome at the equator of the cell. Each of the daughter chromatids contains up to 15 cm (6 inches) of DNA.
But that is still larger than the nucleus by a factor of 5,000 or so (figure 6).
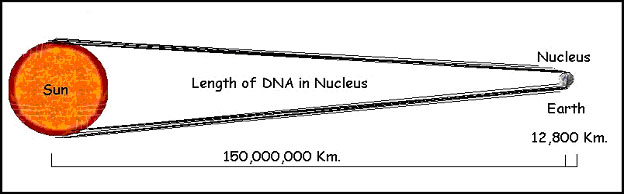
Figure 6. Each cell in our bodies contains approximately 2 meters (6 feet) of DNA. If the nucleus of the cell were the size of the earth, that much DNA would extend to the sun and back eight times!
The DNA must therefore be organized in a very precise way to allow the cell to have access to the needed genes, and at the same time to allow the DNA to be duplicated, and precisely divided to the daughter cells during cell division. This process is facilitated at the most basic level by the association of the DNA with a class of proteins called histones. These very precise proteins come in 5 different forms, referred to as H1, H2a, H2b, H3 and H4. H2a, H2b, H3, and H4 with the help of some associated proteins form a very stable octamer containing two copies of each molecule. Because all of the histones are positively charged to enable them to interact with the negatively charged DNA, the assembly of the octamer requires the aid of several special scaffolding proteins.
This assembled histone core structure is so fundamental to eukaryotic cells that it is preserved across the entire spectrum of living eukaryotic cells with almost no modification. For example, only three amino acid changes distinguish the histone H3 of a pea from that of a sea urchin or a trilobite. Human H4 differs from pea H4 by only two amino acids! This similarity poses other constraints on the problem of origins. If two proteins are that similar across the entire spectrum of organisms, there must be severe constraints on the allowable substitutions. But experimental evidence suggests this is not so, and that viable substitutions can be made at about the same rate as with any other similar sized protein. This begs the whole question on the meaning of similarities, time and biological clocks. What mechanism can explain its origin?
One and a half turns of the DNA molecule (about 140 base pairs) are then wrapped around each histone core to form a nucleosome (figure 7).
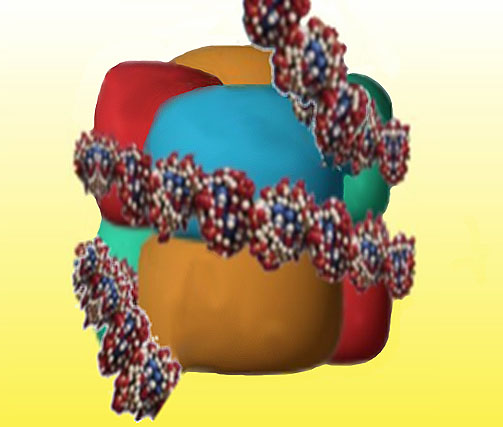
Figure 7. Each Eight of the histone proteins form a cluster that wraps one and a half turns of DNA, greatly reducing the length of the chromosome.
The nucleosomes are associated into larger structures by the binding of the H1 histone. These structures, called solenoids, consist of an array of six nucleosomes in a flattened helix, further shortening the whole molecule. These helical solenoids are then themselves coiled in a complex arrangement that is anchored to the backbone of the chromosome itself . The backbone is composed of a class of topoisomerase proteins with remarkable properties. These topoisomerases (topo II) are connected to the DNA molecule at specific sites. The enzyme can cut one strand of the DNA molecule at the point of attachment, hang on to the cut ends, while passing the uncut strand through the cut ends, then join the two ends of the cut strands again! The resultant structure has accomplished the unfathomable: condensed a molecule of DNA 10 cm long into a structure 50,000 times smaller. But the complexity has only begun.
Each human cell has 46 of these structures that must be duplicated (92) and then correctly segregated so that each daughter cell receives a complete set of 46 chromosomes. Ninety-two separate bodies are moving through the cytoplasm on an unerring journey to the correct daughter cell. The chromosomes contain a special patch of protein called the kinetochore. The attachment of the microtubules to the kinetochore binding region on the chromatids results when a microtubule, engaged in a series of thrusts produced by rapid elongation, contacts a kinetochore of a chromatid and binds to it. If the microtubule fails to contact a kinetochore, it condenses and then thrusts off in a different direction until it has engaged a kinetochore. Once sufficient microtubules from opposite ends of the cell have attached to the two kinetochores of each chromosome pair, the microtubules begin to pull in opposite directions resulting in the equatorial alignment of the chromosomes so familiar in metaphase.
The two chromatids separate at the centromere, and are pulled through the cytoplasm to opposite ends of the dividing cell. The mechanism of movement appears to be the contraction, expansion and depolymerization of microtubules pulling the chromosomes through the cytoplasm in the correct direction. These mechanisms are present in all eukaryotic cells, and the involvement of microtubules and actin and myosin -like proteins in the cell division process illuminates the complexity of a feature that must occur in all eukaryotic cells, including those of trilobites, the first metazoan fossil of record. Keep this in mind while we explore an additional feature of animal cells in particular: the transmission of a nerve impulse.
Directed Protein Synthesis:
Protein synthesis is a subject to inspire admiration, but one we shall not consider here. Rather, taking protein synthesis as a given for the moment, let us examine how proteins are designed to arrive at their correct destination in the cell by directions contained within the protein molecules.
Many proteins function in the cytoplasm where they are produced. These proteins probably need little information targeting them to specific locations. But a large number of proteins must arrive at specific destinations within or outside of the cell in order to function. For example, some proteins are designated to function within the membrane of the endoplasmic reticulum of the cell. Others must be secreted to the outside of the cell, or perhaps in the outer or inner membrane of the mitochondrion , the intramembranous space, or into the mitochondrial matrix. Correct targeting of the protein to each of these different space areas requires explicit instructions within the targeted protein.
In the case of the mitochondrion, an organelle of the cell responsible for converting stored energy into ATP, there are four distinct target areas. Although the mitochondrion has its own DNA and protein synthesizing equipment, most of the mitochondrial proteins are made from DNA contained in the nucleus of the cell. These proteins are produced in the cytoplasm, and must navigate from there into the correct compartment of the mitochondrion (figure 8).
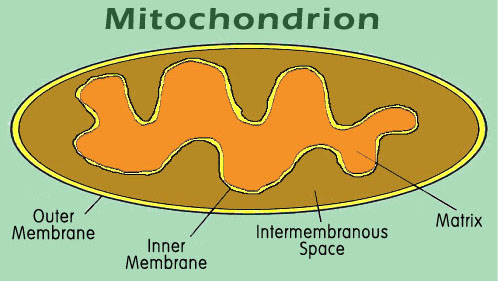
Figure 8. The mitochondrion contains four different target areas that must be separately coded in the protein. The matrix is the site of most of the metabolic activity, and of most of the proteins of the mitochondrion. Both inner and outer membranes are separately targeted by a variety of other proteins, and the intermembrane space is the site for several of the cytochromes.
The targeting processes are complex, and some of the details are still being elaborated, but many of the features are well understood. For example, one possible route for a protein targeted to the mitochondrial matrix is illustrated here. Each compartment of the mitochondrion is handled by a different set of signals and signal receptors, with the result that each protein arrives at the correct destination.
Some proteins must remain within the membrane, either the endoplasmic reticulum, or the outer cell membrane, or in some other cellular membrane. Such proteins play a vital role in regulating the passage of materials through the membrane, and in other vital cellular processes. In order for the protein to be produced in this configuration, the gene for its production must contain, in addition to the usual information on how to build an active functional protein, a variety of instructions informing the cell as to what destination and pathway the protein must follow.
One of the possible destinations for a protein is the outer cellular membrane. Proteins destined for this fate begin with a special set of instructions termed a signal. This system of signals are recognized by a special cytoplasmic body called the signal recognition particle (SRP) (figure 9).

Figure 9. The Signal-Recognition Particle (SRP) is part of a complex of proteins responsible for targeting proteins to specific compartments of the cell. The mechanisms, components and the targeting information appear to be universal, being recognized in plant, animal, and even yeast cells. Proteins destined for targets outside of the cytoplasm (either in membrane-bound compartments, or in the membranes themselves, or for secretion outside the cell), are designated by specific sequences of amino acids in the leader region of the protein. The particle responsible for identifying these specific "signal peptide" sequences, is the SRP. This complex consists of a chain of 300 specific bases of RNA and six proteins, identified by their respective molecular weights (in kilodaltons): P9, P14, P54, P68 and P72. It is known that the P54 protein is responsible for reading and interacting with the signal peptide, the two small proteins interact with the ribosome, and the large P68/P72 proteins are involved in the movement of the nascent peptide chain. The SRP will stop protein synthesis after about 70 amino acid residues, in the absence of suitable membrane interactions, preventing the synthesis of proteins in inappropriate environments.
This particle identifies a coded message in the first 50 or so amino acids of non-cytoplasmic proteins as they are being produced by the ribosome, and binds to this leader sequence, referred to as the "signal peptide". The subsequent steps are detailed in a series of diagrams.
Molecules such as the voltage-gated sodium channel protein discussed below must have encoded within them all of the information for their functional and structural attributes, as well as information for acquiring their active domains, distributed throughout the entire length of the molecule.
This signaling mechanism is universal, since the processes operate in the same way in virtually all eukaryotic cells, including yeast, plant and animal cells. Further, the proteins comprising the translocon and the SRP receptor, that are responsible for the insertion of membrane-targeted proteins into the membrane are also multipass membrane-bound proteins. This means that they also must be inserted into the membrane by a similar mechanism. The universality means the proteins and the mechanism for their acquisition by membranes were already present in the first metazoans of record, and in the ancestral eukaryotc cell, as well.
The Synapse:
The resting neuron or nerve cell has an electrical potential on its membrane of about 60 millivolts negative on the inside. This potential is established by a special sodium/potassium pump which uses cellular energy to pump positively charged sodium ions out of the cell. The nerve impulse is initiated and propagated by an influx of sodium ions into the cell through special protein sodium channels in the membrane.The propagation is mediated by the successive opening of channels in voltage gated sodium channel proteins in the membrane along the length of the axon. This protein makes 24 crossings of the external cellular membrane (figure 10).
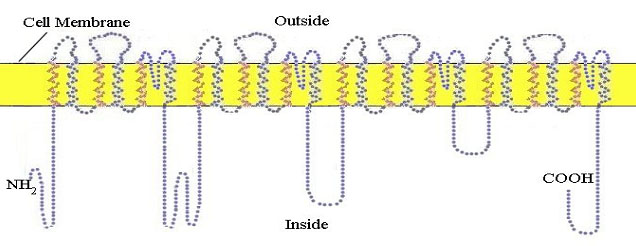
Figure 10. The 24-pass voltage-gated sodium channel protein shown here at completion of its synthesis in the membrane of the endoplasmic reticulum prior to incorporation into the outer cell membrane.
The voltage-gated sodium channel proteins are not only very complex in their construction, as we have seen, but are intricately designed for their functionality in a surprising way. The protein forms a channel through the outer cell membrane that contains a voltage operated gate, and a critical reverse flow guard (figure 11).
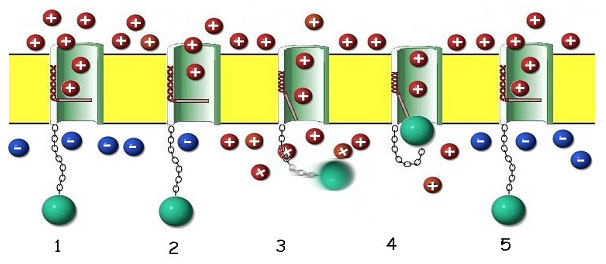
Figure 11. The 24-pass Voltage Gated Sodium Channel Protein as it may appear in its assembled form within the outer cell membrane (yellow). 1. When the membrane is at rest, the channel is blocked by a gating portion of the protein. The resting membrane maintains a positive charge on the outside and a negative charge on the inside. This charge disparity accrues as a consequence of two factors. A sodium-potassium pump (not shown here) forces sodium (red, positive charge) outside the cell membrane, leaving the anionic (blue, negative charge) proteins behind in the cytoplasm. 2. When the sodium channel protein senses a change in voltage resulting from the opening of adjacent pores (depolarization), a solenoid-like portion of the protein in the wall of the channel (red coil) begins to contract. 3. The coil opens the gate as it contracts, allowing sodium ions to rush through the channel into the cell, motivated by diffusion and the potential gradient. The timing of this process is critical. If the channel continues to admit sodium, the cell cannot respond to further stimuli and would quickly die. 4. Since channel cannot close while the membrane is depolarized, a special portion of the protein (green ball) acts like a plug to close the channel until the sodium-potassium pump can restore the resting membrane potential. 5. In a few milliseconds, the pump has removed the excess sodium, the membrane potential is restored, the gate is closed, and the plug relaxes, ready for the next stimulus.
As the depolarization of the nerve is sensed by the pore protein, the gate opens and sodium ions flow into the cytoplasm, propagating the voltage change and triggering the same response in adjacent pores. Once the membrane is fully depolarized, the special blocking segment of the pore protein plugs the channel, preventing further depolarization until the resting membrane potential has been reestablished by the sodium ion pump. When a nerve impulse reaches the terminus of a nerve, it must pass the signal across a gap to the next nerve cell. The juncture of the two cells is called a synapse, and the gap separating the two cells is called a synaptic cleft (figure 12).
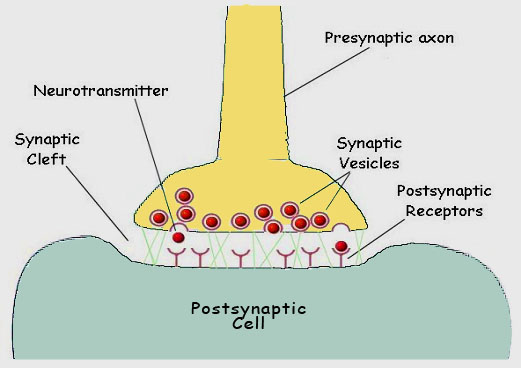
Figure 12. The chemical synapse. When one nerve must pass an impulse on to the next cell, it does so by one of two general processes. Where speed is the critical factor, a direct electrical connection may exist between the two cells Where modulation is important, a chemical event intercedes between the arrival of the impulse at the terminal of one axon and the continuation of the signal in the next cell. The juncture is referred to as a synapse. The cells are separated by a gap, the synaptic cleft. Upon appropriate stimulation the presynaptic cell releases neurotransmitter (commonly acetylcholine). The neurotransmitter then diffuses across the cleft to receptors on the postsynaptic membrane. Here the receptors respond by initiating depolarization of the membrane and the impulse is propagated in the postsynaptic cell.
In many cells the transmission is mediated by the release of a neurotransmitter substance, often acetylcholine, a small biomolecule. The acetylcholine is accumulated in cytoplasmic membrane vesicles, where a hydrogen ion antiport protein exchanges acetylcholine, made in the cytoplasm of the cell, for hydrogen ions, pumped into the vesicle at the expense of energy from ATP hydrolysis. The vesicle is then transported through the cytoplasm along the microtubules of the cytoskeleton towards the membrane of the synaptic surface (figure 13).
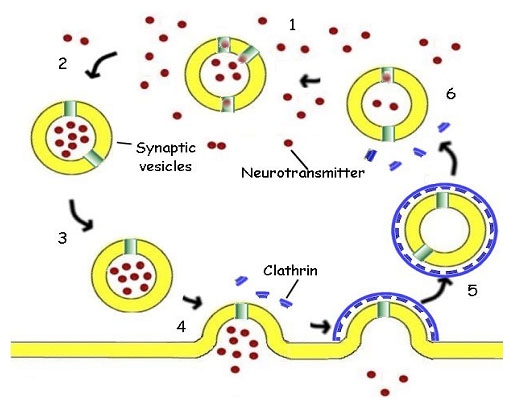
Figure 13. The Synaptic Vesicle. 1. The synaptic vesicle accumulates hydrogen ions at the expense of ATP. The hydrogen ions are then exchanged for acetylcholine (red), a neurotransmitter, by a specific antiport protein. 2. As the vesicle becomes charged with acetylcholine, it is picked up by a cytoplasmic transport protein and carried through the cytoplasm to the synaptic region of the neuron. 3. The filled vesicle is docked in the region of the synapse, where it awaits a nerve stimulation. 4. Triggered by an influx of calcium ions, the vesicle releases its charge of acetylcholine to the synapse, passing the impulse on to the post-synaptic cell. 5. The empty vesicular membrane is surrounded by cytoplasmic protein molecules called clathrin (blue), capturing the vesicle for reuse. The clathrin forms a cage coating the entire vesicle. The vesicle then travels back away from the membrane into the cytoplasm and loses its protective cage. 6. The vesicle again begins to accumulate hydrogen ions the cycle is repeated.
This process of moving the vesicle through the cytoplasm is itself amazing, as kinesin-type molecules have two legs that simply walk along the elements of the cytoskeleton in a very anthropomorphic fashion, carrying the synaptic vesicle along with them.
In the membrane of the synaptic vesicle are a number of unique proteins not found elsewhere in the cell membrane. Two of these are synaptobrevin and synaptotagmin. Synaptobrevin binds a complex of proteins called NSF (N-ethylmaleimide Sensitive Factor) and SNAPs (soluble NSF associating proteins). This complex binds to syntaxin, a protein found only in the plasma membrane in the region of the synapse, thus anchoring the vesicle to the membrane. Synaptotagmin, the other mentioned vesicle protein, has two binding sites for Ca++ on its cytoplasmic side. In the absence of Ca++, synaptotagmin binds to the synaptotagmin-syntaxin-NSF-SNAP complex, and prevents the binding of alpha SNAP, the fusion protein. When a nerve impulse reaches the synaptic region, calcium channels are opened, admitting Ca++ to the cytoplasm. Synaptotagmin binds the calcium and alpha SNAP can then bind to the complex. As a result, the membrane of the vesicle fuses with the cell membrane by a mechanism not yet resolved, expelling the contents of the vesicle into the synapse, and triggering the response of the neighboring cell.
Also involved in the sorting and dispersal of the cytoplasmic vesicles are Rab proteins, which are the UPS system of the cell. These proteins are attached as shipping labels to all of the various vesicles in the cytoplasm to specify their ultimate destination. When they arrive at their destination, the shipping label is read and if the destination is that specified, the vesicles are permitted to fuse and share their contents with the recipient organelle. If the label specifies some other destination, the vesicle is refused access to the organelle.
Meanwhile, cytoplasmic proteins called clathrin identify the depleted vesicle, and surround it with a hexameric cage that preserves the membrane and the associated proteins from being lost. The clathrin cage remains attached until the vesicle can be reunited with its host endosome in the cytoplasm for refilling. This process which I have described in just the barest details, is common to all animals with nervous systems, from the simplest invertebrates to man. Because this process represents a very complex mechanism shared by insects and humans, we can be absolutely confident trilobites worked this way too.
The Developmental Biology of Trilobites
What can we say about the complex pathways by which a single ovum in the ovary of a mother trilobite became a functioning offspring: a great deal more than you might imagine thanks to recent advances in our understanding of molecular biology of development. Here I will only be able to give the sketchiest details. Let me quickly tell you a little about how an insect is formed. Here we will discuss a metamorphosing insect, the fruit fly Drosophila.
Because these insects are quite small, it would be impractical to hatch a fully functional winged offspring from a single fertilized ovum. The strategy of many insects is to lay an egg, which then "hatches into a bigger egg, called a caterpillar. The caterpillar is just a bag for accumulating food material in preparation for the production of the adult form. However, deep within the recesses of each caterpillar are the embryonic seeds of an entire adult organism. Termed "imaginal disks", these specialized tissues remain dormant until pupation, at which time the caterpillar dissolves, and the imaginal disks become the various parts of the adult. This is in itself an amazing process, but the sequence of events leading up to the formation of the imaginal disks give an unprecedented view into the process of development that will be of great interest to us in our consideration of the trilobite.
While the egg is still within the ovary, gradients of specific regulatory gene products are established within the egg. These mRNAs or proteins originate either from the egg nucleus itself or from maternal accessory cells surrounding the egg in the ovary. Subsequent to fertilization additional series of genes are activated, producing additional regulatory proteins in specific regions of the fertilized egg. This asymmetric distribution of regulatory proteins results in each cell having a unique combination of regulators. The balance of these gene regulators determine which genes are activated and which are suppressed in each cell and this asymmetry in turn determines head and tail, and differentiation along the resulting body axis.
This whole system of development is fantastically complex. Genetic studies in Drosophila revealed a class of developmental genes which when mutated resulted not just in a single change, such as eye color, but produced either massive effects which were lethal, or resulted in changes in body form on a monstrous scale. For example a single gene mutation in one of the regulatory genes resulted in legs growing where the antenna are normally found, or in the formation of an extra body segment with an extra set of wings. Vast regulatory networks link each of these developmental genes to hundreds of other genes.
Much to their astonishment, the investigators found the genes that were controlling development of fruit flies and the genes that controlled the development of vertebrates, including mice and men, were very similar in structures, and that the genes often controlled analogous parts of the embryos of flies and men. And thus, these developmental gene sequences, present in flies and men, were also present in trilobites.
Subsequent study revealed the location of some of these genes on the chromosome. When the major series of regulator genes that determine the polarity of the Drosophila embryo (HOM-C genes) were identified and mapped, the investigators discovered an amazing fact; one they were not expecting and were not equipped to deal with when they found it: The genes that controlled development of the axis of the embryo from head to tail lay on the chromosome in the same order as the portions of the anatomy of the organisms whose development they were intended to control (colinearity). That is unexpected for a variety of reasons, not the least of which is the improbability of such an arrangement occurring in the absence of a designer.
Some years ago, Murry Eden, a mathematician from M.I.T., demonstrated the improbability of obtaining genes in a specified order on the chromosome. There appears to be no functional reason for them to be so ordered, although this picture could change. But that was not the most astonishing thing. Subsequent studies on vertebrates (mostly on mouse, but also on human), revealed similar types of regulatory proteins was responsible for ordering the head to tail organization of the body of vertebrates, including man. And these genes (called Hox genes) which were very similar to the equivalent genes in Drosophila, (for some homeotic genes the similarity between Drosophila and human is 98%) lay on the chromosome in the same order as those in the fruit fly!
They must have had a common origin! And they must have been present in the trilobite, one of the earliest metazoan forms of the Cambrian. Thus not only were all the complexities of the eukaryotic cell present in the first form, but all of the unfathomable complexity of the system of development, involving the interaction of thousands of genes, that all cephalized forms appear to have in common, were in place in these organisms.
The Trilobite Eye
The eye has been an object of wonder throughout recorded history because of its critical functions. Surely the existence of fully functional compound eyes on this early metazoan has from time to time caused thoughtful evolutionists to seriously question the basis of origin. In the case of trilobites, not only were these early appearing forms equipped with highly organized visual organs, but some of the recently discovered properties of trilobite eyes represent an "all time feat of function optimization." The trilobite eye (figure 14),
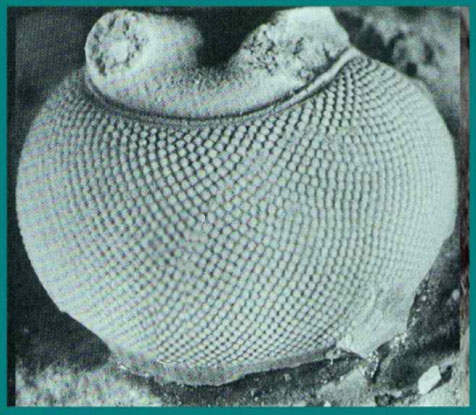
Figure 14. A trilobite eye illustrating the complex compound structure.
from what we can gather by study of the fossil forms, shares much in common with modern insect eyes (figure 15).
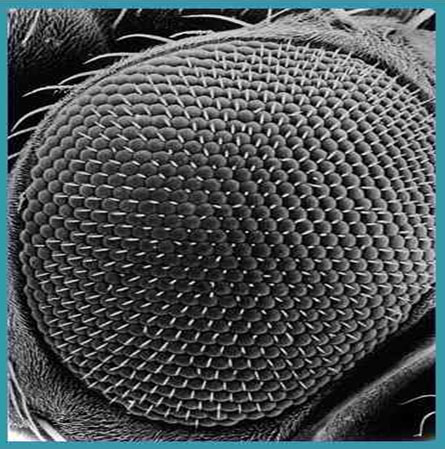
Figure 15. . The eye of a modern insect illustrating a compound structure similar to that of the trilobite.
Certain trilobites of the Early and Middle Paleozoic have a unique optical system unknown in any other creature. The nuclear physicist (Director of the Fermilab at U. Chicago) and trilobite aficionado Levi-Setti states with unabashed candor:
"And a final discovery---that the refracting interface between the two lens elements in a trilobite’s eye was designed [emphasis added] in accordance with optical constructions worked out by Descartes and Huygens in the mid-seventeenth century--borders on sheer science fiction."
The axes of the individual ommatidia were constructed of single crystals of calcite with the optical axis of the crystal coincident with the optical axis of the eye element. That presents an unusual problem for the trilobite, since a simple thick spherical lens of calcite could not have resolved the light into an image (figure 16).
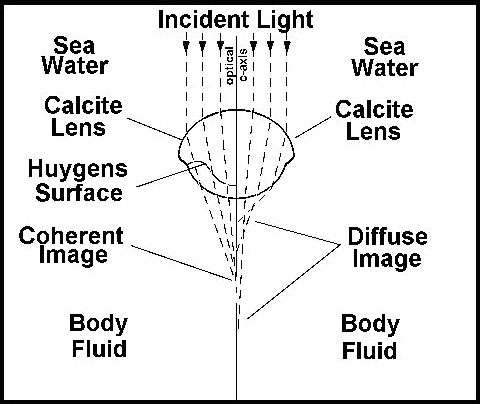
The trilobite optical element is a compound lens composed of two lenses of differing refractive indices joined along a Huygens surface. In order for such an eye to correctly focus light on the receptors it would have to have exactly this shape of lens. The optical principles required were first elaborated by Huygens in the 17th century, but the trilobite lens worked perfectly using these optical principles long before the Dutch mathematician figured out how. Now the earliest trilobites lacked these sophisticated lenses, but had eyes that were apparently more like those of modern insects. But no intermediate forms are known from the fossil record. When the Huygens lens first appears, it is fully functional.
The regulatory mechanism of eye development must indeed be complex. An estimated 2500 - 5000 genes are involved in the developmental process. Some of the details of development are again being worked out in Drosophila where some of the master switching genes are known. The individual facet, or ommatidium of a compound eye such as that in Drosophila, consists of a cluster of eight cells, seven of which will develop into light receptors. One of these retinal cells, called R7, was found to be responsible for detecting UV light. The developmental pathway of R7 has been the subject of intensive investigation for a number of years. It has revealed a cascade of interactions that seems to typify most externally binding ligand pathways in cells.
The membrane of the R7 cell contains a special protein, the Receptor Tyrosine Kinase (RTK). This protein includes an extracellular receptor site, a transmembrane portion, and an intracellular enzymatic portion. When an external ligand becomes bound to the receptor (in this case it is a membrane bound ligand on the eighth cell), the molecule joins with another RTK, forming a dimer. The two molecules then engage in reciprocal phosphorylation of three specific tyrosine residues, each on the other molecule.
Thus phosphorylated, the cytosolic portion can bind a specific cytoplasmic protein (GRB2) that recognizes the phosphorylated RTK. When GRB2 is bound to RTK, it can then bind a third protein, Sos (Son of sevenless). The Sos complex causes the membrane-associated protein, Ras to lose GDP, which is then replaced by GTP. In this condition the Ras protein binds a protein called Raf, a threonine/serine kinase. When bound by activated Ras, Raf is able to bind and phosphorylate and thus activate another specific tyrosine/threonine kinase, MEK. MEK in turn activates a cytoplasmic enzyme, MAP Kinase, by phosphorylating tyrosine and threonine residues on this enzyme.
MAP Kinase is apparently involved in phosphorylating DNA binding proteins and other key cellular proteins that result in changing the direction of cellular differentiation, so that such a cell will now become a normal R7. What is especially noteworthy about this cascade, is that it is found in all multicellular eukaryotic cells, and with slight differences in the single celled eukaryotes as well (yeast and protozoa).
Recently as a result of manipulations of a master developmental gene, eyeless, flies have been produced with eyes appearing on various parts of the body, including the wings, legs and tips of the antennae as a result of activating the gene in unnatural positions. A similar master gene has been found in vertebrates, which have eyes completely different from those of insects. The gene in humans, mice and other organisms is nearly identical with that in Drosophila. When the appropriate gene from a mouse chromosome (and presumably from a human as well) was inserted into a fly, it produced fly eyes wherever it was activated on the body of the fly! The two genes are similar enough that the mammal gene can cause the formation of an insect eye. Our line of reasoning leads us to conclude that the system of genes leading to development of the eye was present and functioning in the first trilobites.
There are a rapidly increasing number of developmental pathways that are "evolutionarily conserved," a euphemism for "embarrassingly similar," across a broad spectrum of organisms and most of these would have been present in the trilobite . For example, the genes responsible for the organization of the dorsal-ventrality of the human were discovered using the genes from drosophila as molecular probes. The genes responsible for the organization of the human brain in embryogenesis were discovered using the genes from Drosophila as probes. The eye, the hindbrain and spinal cord, the pathing of axons, the differentiation of skeletal and heart muscle, the photoperiodic response, the sculpting of tissues involving select cell death (apoptosis), embryonic patterning, cell signaling, and a thousand other examples of "evolutionarily conserved" processes could be cited. Even the formation of limbs is directed in fruit flies by a gene (Hedgehog), whose homologous gene in vertebrates (Sonic Hedgehog) directs the formation of limbs in all known vertebrates, including, human, mouse, chick and even fish! The elaborate mechanism responsible clearly precedes any known organism with limbs. Where did all of this information come from?
The Problem of Complexity for the First Metazoa
I have just presented a few brief examples illustrating the complexity of living eukaryotic cells and organisms. These were drawn from among hundreds or thousands of other examples that could equally well have been used, in order to make the following points. One of the first complex animals of which we have a knowledgeable record, the trilobites, appeared in the early Cambrian. The basal Cambrian is sometimes loosely defined as the point in the geologic column where the first trilobites appear.
Trilobites are arthropods, in the same alliance as modern insects. The cells of trilobites divided in a manner similar to every modern eukaryote. The mechanisms were all in place, all functioning as they do today. The trilobites had nervous systems as complex as those of modern insects. The synapses in the nervous systems of trilobites functioned just as the synapses of all modern organisms do. The eyes of trilobites manifest all of the complexity and developmental integrity of modern forms. The complexities that I have just described, were all present, all fully functional in the FIRST MULTICELLULAR ANIMALS FOR WHICH WE HAVE RECORD. WHERE AND WHEN DID THE EVOLUTION TAKE PLACE???
This question has been adroitly avoided by evolutionists. The systems we have just described did not happen by accident. They were designed. Every step taken by a trilobite was an indictment of the inadequacies of evolutionary theory. This is why, when evolutionists such as Steven Gould write books about the earliest life forms, they carefully avoid mention of the problem of infinitely complex forms suddenly appearing. Their attitudes are "Its there, therefore evolution must be able to do it." Evolution is a black box. It is magic! It is EVOLUTION!
Concerning the dearth of fossil evidence in the Precambrian, Leonard Brand (1998) has written:
"One of their [evolutionary scientists] biggest assumptions was that the molecular clock is reliable.... When Levinton gave his paper [at the 1996 GSA meetings in New Orleans] he stated that the molecular clock can be best compared to a sun dial in the shade, which isn’t very encouraging for his method, but he and his colleagues still believed that it yielded data sufficient to test the theory of the rapid evolution of life at the base of the Cambrian... .
From their molecular clock data they concluded that the initial divergence of metazoan life forms occurred about 1.2 billion years ago (+/- 50 to 250 million years) . The base of the Cambrian is currently dated at about 543 million years ago , so their conclusions require a half billion years of metazoan history before the Cambrian. They also concluded that the beginning of Metazoan phyla was not an explosion, but was somewhat spread out during that half billion years.
A couple of days later these papers were discussed in a 'Hot topics discussion' during the noon hour. Four scientists gave brief presentations on the new ideas about the Cambrian explosion, followed by audience questions and comments. Many questions dealt with technicalities of their research method, but two questions stand out. A little background is necessary before dealing with these questions. The proposal that complex metazoan animals, ancestral to such things as molluscs, trilobites, vertebrates, sea urchins, corals, and many others, existed for a half billion years before the Cambrian implies that they lived all that time without leaving a fossil record. This pretty much requires that before the Cambrian they existed as soft worm- or larvae-like forms, with the general genetic make-up of the Cambrian groups but without their skeletonized morphology.
Now the questions. The first of the two questions was - why are trace fossils (fossil tracks, trails, and burrows) so rare before the base of the Cambrian, if these animals existed for that half billion years? An internationally recognized expert on trace fossils stood up, presumably to answer the question. However, he talked about other things and the very important question never was answered. At the end of the discussion another scientist stood up and commented on the implication that all the skeletonized phyla developed skeletons at about the same time in the Cambrian. He asked - why are all these types of animals living for so long and then all making skeletons all at once? He then asked, with some vigor - 'Why are you avoiding the real question?' After a pause, one member of the original presenters answered 'because it’s really hard (a hard question)'. He went on to say that they hoped answers would come from further study of developmental biology.
These two questions were apparently not asked by people who doubted the evolution theory, but by evolutionary scientists willing to ask the hard questions that need to be addressed as they try to test between different hypotheses. The fact remains that the Cambrian explosion is one of the big challenges to naturalistic theories that still remains unanswered."
Some recent quotations from an article in Time magazine (When life exploded, TIME, 12/5/95) on the origin of the Precambrian metazoa are instructive in helping us to realize just how close some evolutionists are coming to truth: The article pointed out that all animal phyla except perhaps bryozoa are present in early Cambrian, and that they all appear within a very small slice of time ("no more than 10 million years").
Steven Gould of Harvard (paleontologist): "Fast is now a lot faster than we thought, and thatís extraordinarily interesting."
Samuel a Bowring, M.I.T. (geologist): "We now know how fast fast is, and what I like to ask my biologist friends is, How fast can evolution get before they start feeling uncomfortable?"
Rudolph Raff, Indiana U. (biologist): "There must be limits to change. After all weíve had these same old body plans for half a billion years."
G. M. Narbonne, Queens U. (paleontologist): "What Darwin described in the Origin of Species was the steady background kind of evolution. But there also seems to be a non-Darwinian kind of evolution that functions over extremely short time periods--and thatís where all the action is."
Conclusions:
We have given careful consideration to a small sampling of the thousands of examples of shared complexities of modern forms that could have been used. We have seen that from a careful consideration of the evidence, evolutionary theory does not explain the origin of the information-rich systems of biological organisms.
We have seen that the first abundant, well represented metazoan fossils, the trilobites, were complex beyond imagination in every detail, with compound eyes, with swimmerets and gills, with legs and antennae, and with complex, even intricately sculpted forms. They had fully functional muscular and nervous systems. Their eyes were developed by processes not only similar to those of other arthropods, but like those of vertebrates, including man. The complex system of development of cephalized forms was already present and functioning. A thousand other complexities of molecular biology shared by modern forms were operative. Where did these complexities come from? There is no evidence of any earlier form from which they could have been derived.
The trilobite, among the first metazoan forms found in the Lower Cambrian, was FULLY as complex at the molecular level as any modern form. Furthermore, there is no support for a mechanism in biological systems for adding information to complex systems (Spetner, 1998). To argue that they came from Precambrian forms that were not preserved because they had no hard parts is to argue again from the ABSENCE OF EVIDENCE.
The absence of evidence, in science has to be construed as the evidence of absence. There is no Precambrian evolutionary sequence because there was no Precambrian evolution. Evolution as an explanation for the existence of complex living systems is a religious view held by those who wish the world to have no Originator (meaning – Huxley 1937, p. 312). Trilobites and all other forms appear on the scene as fully formed, fully competent organisms, period. It is past time to replace the theory of organic evolution with a theory that can explain the data. The only theory with explanatory value for the origin of information is the theory of Special Creation. I make no apology for choosing to place my faith in the existence of a Master Designer, a position that is consistent with the clearest interpretation of the evidence available in the Geologic Record, consistent with the clearest reading of the Book of Genesis, and a faith that is positive, uplifting and full of hope for the future.
References
Brand, L. 1998. Personal communication.
Lodish, Harvey, et. al. 1995. Molecular Cell Biology. Third edition. Scientific American Books. W. H. Freeman and Co. New York.
Dobzhansky, Th., F. Ayala, G. L. Stebbins and J. W. Valentine. Evolution. 1977. (W. H. Freeman &Co.San Francisco). p.265, ff.
Wray, G. A., Levinton, J. S. & Shapiro, L. H. 1996, Molecular evidence for deep pre-Cambrian divergences among metazoan phyla. Science, 274, 568-573.
Carroll, Sean. 1995. "When Life Exploded." Time Magazine. 146:(23). Boncinelli, E., Simeone, A., La Volpe, A., Faiella, H., Acampora, D., & Scotto, L. 1985. Human cDNA clones containing homeobox sequences. Cold Spring Harb. Symp. Quant. Biol., 50, 301-306.
Levi-Setti, R. Trilobites. 1993. (University of Chicago Press, Chicago). p. 29. Ibid, p.54.
Rubin, Gerry, Developmental Geneticist U.C. Berkeley. Quoted in "Secrets in a fly’s eye", Discover 17: (7) 110.
Levinton, J. S., G. Wray, and L. Shapiro. 1996. Molecular evidence for a deep Precambrian divergence of animal phyla. I. Introduction and regression approach. Abstracts with Programs, 28 (7): A-52. Geological Society of America annual meeting, Denver, CO.
Wray, G., J. S. Levinton, and L. Shapiro. 1996. Molecular evidence for a deep Precambrian divergence of animal phyla. II. Relative rate tests and implications. Abstracts with Programs, 28 (7): A-52. Geological Society of America annual meeting, Denver, CO.
Grotzinger, J. P., S. A. Bowring, B. Z. Saylor, and A. J. Kaufman. 1995. Biostratigraphic and geochronologic constraints on early animal evolution. Science 270:598-604.
Huxley, Aldous. 1937. Ends and Means.
Spencer, Lee A. 1998. Not by Chance! Judaica Press.
